What is epitranscriptomic modification?
RNA modifications, not limited to purine or pyrimidine bases, including N6-methyladenosine (m6A), 1-methyladenosine (m1A), 5-methylcytidine (m5C), 5-hydroxymethylcytidine (hm5C), 5-formylcytidine (f5C), 5-carboxycytidine (ca5C), inosine (I), pseudouridine (Ψ), and 2′-O-methylation (Nm). RNA modifications can structurally alter the pairing of nucleobases, leading to structural rearrangements of RNA and thus regulating the function of the molecule. Epistatic transcriptome modifications determine the multifunctional nature of RNA and the large number of biological processes it regulates, including RNA splicing, translation, cellular localization and lifespan. Therefore, decoding information about modifications that affect sequence or structural changes is increasingly important.
Methods for detecting RNA modifications
Currently, most methods used to detect RNA modifications fall into three categories: high-throughput whole-transcriptome sequencing technologies, mass spectrometry methods, and bioinformatics analysis. While the advent of next-generation sequencing provides a wealth of sequence information in a single run, it requires advanced algorithms to process and interpret this information. To improve sensitivity and obtain more comprehensive data, next-generation sequencing has been combined with immunoprecipitation (IP) and base-specific chemistry. In addition to various high-throughput sequencing techniques, mass spectrometry (MS) methods have been developed for the identification of nucleic acid modifications. However, this article focuses on sequencing-based assays.
N6-methyladenosine (m6A)
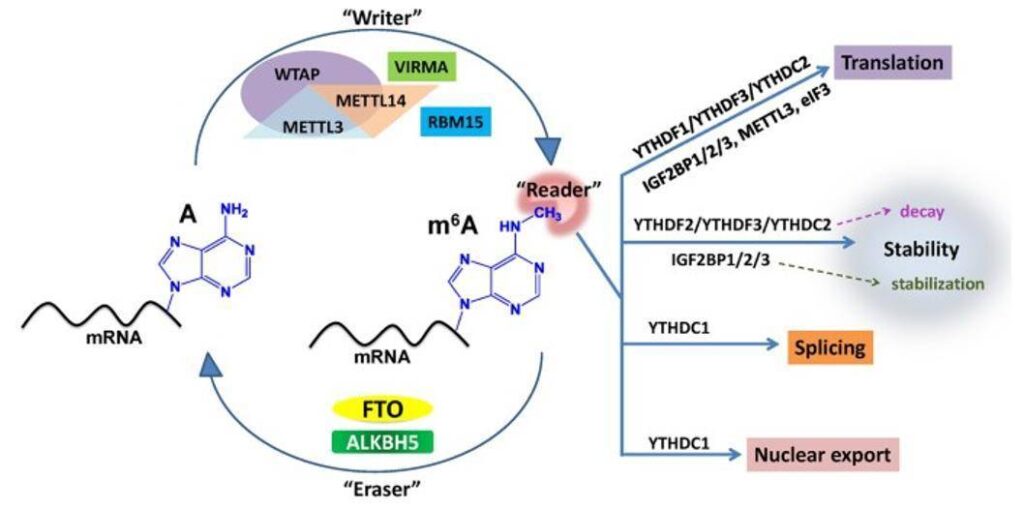
The m6A modification is formed by methylation of adenine at the N6 position and is estimated to occur at a frequency of 0.1-0.6% m6A/A. Most m6A modifications preferentially occur near the stop codon and the 3′ untranslated region (UTR), particularly upstream of the stop codon. m6A is found to be a reversible RNA modification with a group of enzymes that function as “writers” and “erasers”.

Summary of m6A modification machinery in cancers. (Deng X et al., 2018)
Most m6A recognition ways are based on m6A-seq and MeRIP-seq. Enrichment of m6A-specific methylated RNA is performed by immunoprecipitation using m6A-specific antibodies after an initial mRNA fragmentation step, whereas RIP or RNA immunoprecipitation is a variant of IP that targets RNA modifications rather than proteins. Once the target RNA has been enriched, the process continues along traditional RNA sequencing methods.
1-methyladenosine (m1A)
Methylation of the adenine N1 position is approximately ten times more abundant than m6A, with an estimated frequency of 0.015-0.054% m1A/A. This modification occurs predominantly in the GC-rich region of the 5′ UTR and has been identified in tRNA, rRNA and, more recently, mRNA. Similar to m6A, the m1A has a highly dynamic regulatory function. The localization of the m1A modification near the translation start site and the first splice site in coding transcripts was found to correlate with translation modulation. Thus, it can affect translation by causing changes in RNA folding, thus allowing access to previously paired RNA regions. It is also associated with an increase in translation and changes in RNA cell metabolism.
The identification of m1A is similar to that of m6A and can rely on m1A-specific antibodies to enrich for modified RNA, which then proceeds to the sequencing step.
5-methylcytosine (m5C)
Methylation of cytosine at 5th position has been shown to be present in tRNA, rRNA and mRNA. This modification is relatively common, with RNA sequencing showing coding and non-coding mRNA regions with over 8000 m 5 C sites at an estimated frequency of 0.025-0.095% m 5 C/C. The subtle enrichment of m 5 C is evident in both the 5′ and 3′ UTRs, and the distribution of modified bases within this region further favors the Argonaute (AGO) The distribution of modified bases in this region further favors the binding sites of Argonaute (AGO) proteins, which are involved in the RNA interference (RNAi) pathway that inhibits gene expression. Although m5C modification does not interrupt base pairing, it does increase the hydrophobicity of the RNA homeotic groove and may increase base accumulation. Thus, m5C has a stabilizing effect on the secondary structure of tRNA and can also affect the translation fidelity of rRNA. m5C variants, such as hm5C, f 5 C and ca5C, have also been shown to affect the structure of surrounding RNA. In particular, hm5C is present in a variety of transcripts involved in basic cellular processes and development.

High-throughput sequencing technologies reveal that RNA 5-methylcytosine plays dynamic regulatory roles in the diverse cellular processes via its writer, eraser, and reader. (Chen Y S et al., 2021)
Similar to the m6A and m1A detection, m5C RNA immunoprecipitation (m5C-RIP) enriches fragmented RNA with m5C-specific antibodies prior to cDNA library construction and sequencing, whereas BS-seq does not allow differentiation between hm5C and m5C, revealing only the presence of cytosine modifications in RNA transcripts.
The future of RNA modifications detection
A variety of transcriptome-wide techniques are available for detecting modifications to various RNAs. The ideal method for quantitative detection should include single-base resolution and a high level of accuracy and precision. Therefore, to obtain a true picture of the RNA epitope transcriptome, single-cell RNA modification sequencing and more simultaneous multi-modification sequencing techniques are available, as different RNA modifications have the potential to interact within the cellular system. Methylation modifications can currently be identified by single-cell sequencing techniques. Continued advances in epitranscriptomic identification technologies will allow further exploration of how epitranscriptomic modifications affect the structure of RNA and interactions with other biomolecules.
References:
- Van Horn M L, Kietrys A M. Epitranscriptomic modifications and how to find them. Epitranscriptomics, 2021: 165-196.
Read Other: Respite Care
CD Genomics is a leading global life sciences company, and we remain committed to providing the research community with high-quality long-read sequencing services, from Oxford Nanopore to PacBio SMRT sequencing.